|
Getting Started
The aim of this section is to
provide a short tutorial on how to getting started with the RBN
toolbox. This tutorial is rather an overview over some basic
features of the toolbox than an exhaustive explanation of all
available functions. If you want to know what you can do with the
RBN toolbox without reading the whole function
reference, then this is your starting point. If you have
doubts about what Random Boolean Networks are, then have a look at
the RBN Overview.
Initializing and
visualizing the network
First of all, you have to decide
what parameters your Random Boolean Network (RBN) will have. For
reasons of clarity, we deal in this tutorial only with two
parameters: N, the number of nodes in the network and K, the number
of incoming connections per node.
These parameters remain generally constant over time, but there
are also cases where one wants to change the network's topology
after initialization. For this case, please consult the function
reference.
There are a handful
of functions that can be called to set up a Random Boolean
Network. However, if one deals with a "standard"
situation where only the parameters N and K are specified, then
the easiest way to create the corresponding RBN is to use the
bsn()
function (Build and Show Network). In fact, this function calls all the basic initializing
functions with some standard parameters and also calls a function
to visualize the network's topology (displayTopology()).
For example let's create a network with N = 8 and K = 2. At the
Matlab command line type:
>> [node, conn, rule] = bsn(8,2,'arrow');
The variables node, conn
and rule are matrices and contain information about node
states, the logic transition rules associated to the nodes and the
connections between the nodes. These three matrices are necessary
and sufficient to represent the whole network and are therefore at
the core of each session with the RBN toolbox. For details about
internal representation of the network and these three matrices,
have a look at the Matrix
Representation section.
Matlab not only creates the
matrices node, conn and rule, but also
opens a new figure window displaying the network's topology.
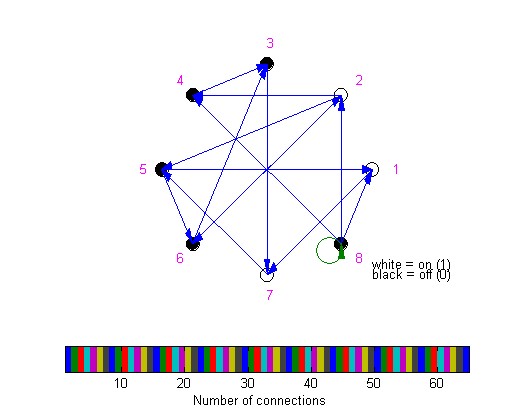
All nodes lie on an immaginary
circle and are represented as small circles colored in black or
white, according to their node state (zero or one). The colours of
the directed connections represent the number of connections
between two nodes, which can be determined by inspection of the
Matlab color bar.
Calculating and
visualizing the network's evolution
Now, we want to evolve our
network over, let's say, 20 discrete time-steps and display the
evolution. In order to do that, we just have to call the displayEvolution() function with the desired update scheme. There
are a lot of different update schemes that are classified by their
degree of synchronicity and determinism. For a description of all
update modes consult the RBN
Overview section. Here we choose for example the
"Deterministic Generalized Asynchronous RBN" update mode
(DGARBN).
>> [node, tsm] = displayEvolution(node,20,'DGARBN');
node now contains information about all
nodes at the end of the evolution, that is after time-step number
20. tsm is a time-state-matrix that contains all states of the
nodes over the evolution time of 20 time-steps. The
displayEvolution() function implicitly visualizes the time-state
matrix:

The x-axis corresponds to discrete time steps and
the y-axis to the nodes. So in our case the time-state matrix is
represented as a grid with 8x21 cells. The first column
corresponds to the initial state of the network, the second column
to the network's state after one time-step, etc.
As the displayEvolution() function returns all information of the
network at the end of the "DGARBN"-evolution, it is
possible to take these node-states as initial values for an other
evolution, let's say, in "Classical RBN" update mode.
(CRBN)
>> [node, tsm] = displayEvolution(node,20,'CRBN');
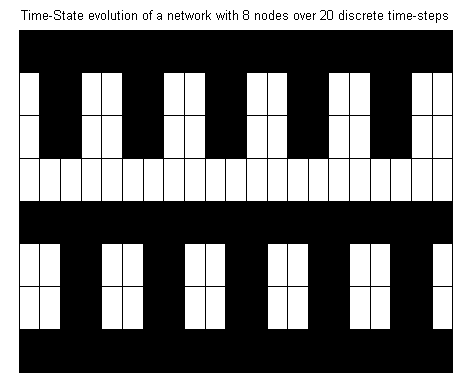
Finding the attractor
Obviously, after some time-steps our time-state
matrix starts to exhibit some regular structure. To find the attractor, its length and the states in it, call the
findAttractor() function.
>> [attrLength, attrStates] = findAttractor(node,tsm);
The variables attrLength and
attrStates now contain the length of the attractor and all states
in the attractor, respectively. By plotting the attrStates matrix,
we find the attractor of our network, which is a sample of the
evolution calculated before.
>> displayTimeStateMatrix(attrStates);
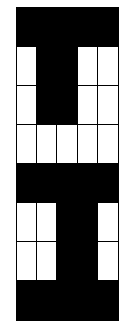
Visualizing node statistics
In this simple example, it
is easy to keep an overview of the dynamics of the network. Yet,
if a network with 30 nodes and 5 incoming connections per node is
being evolved over 100 discrete time-steps in "CRBN"
update mode, it may be useful to
use some statistics.
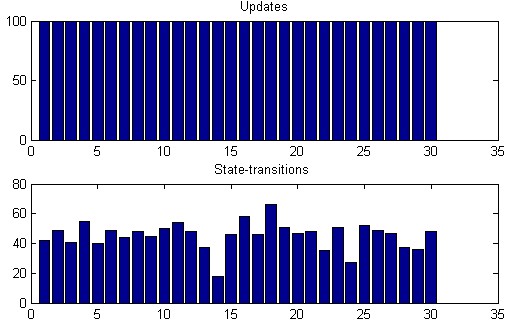
The displayNodeStats() function provides histogramms summing up
the dynamics of the network.
>> [node2, conn2, rule2] = bsn(30,5,'arrow');
>> [node2, tsm2] = displayEvolution(node2,100,'CRBN');
>> displayNodeStats(node2, tsm2);

The corresponding node-statistics show the number
of updates of each node and the effective number of transitions of
each node. As the evolution has taken place in "CRBN"
update-mode, it is clear that each node has been updated at each
time-step.
|